library(tidyverse)02 — Getting Oriented
January 23, 2024
Motivation
Technical computing is often frustrating
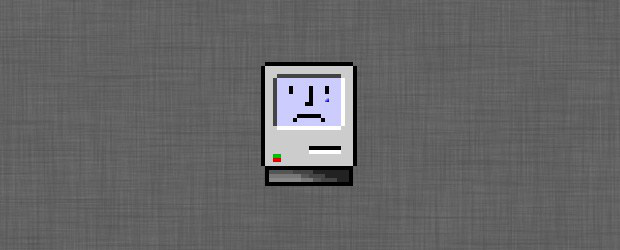
What is this?
Two Revolutions in Computing
What everyday computing is now
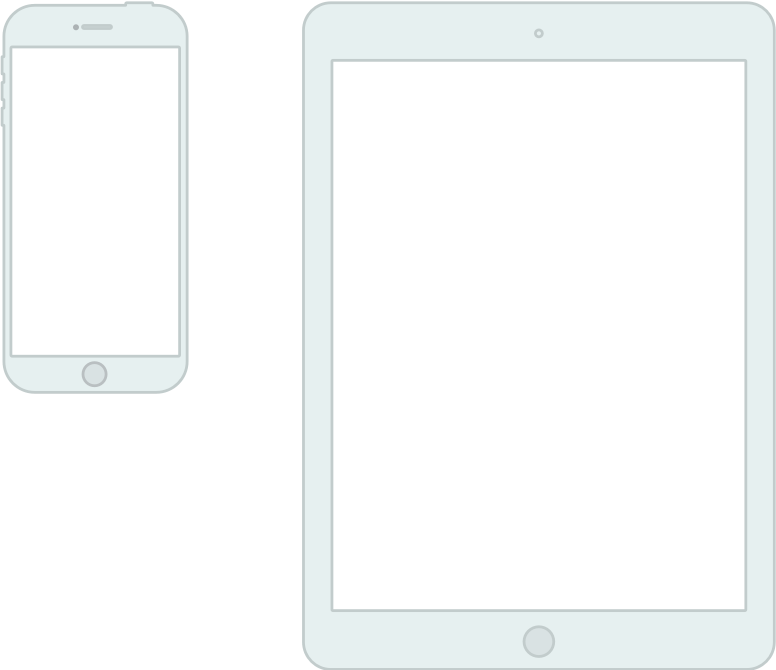
Touch-based user interface
Foregrounds a single application
Dislikes multi-tasking
Hides the file system
“Laundry Pile” user model of where things are stored
Where technical computing lives
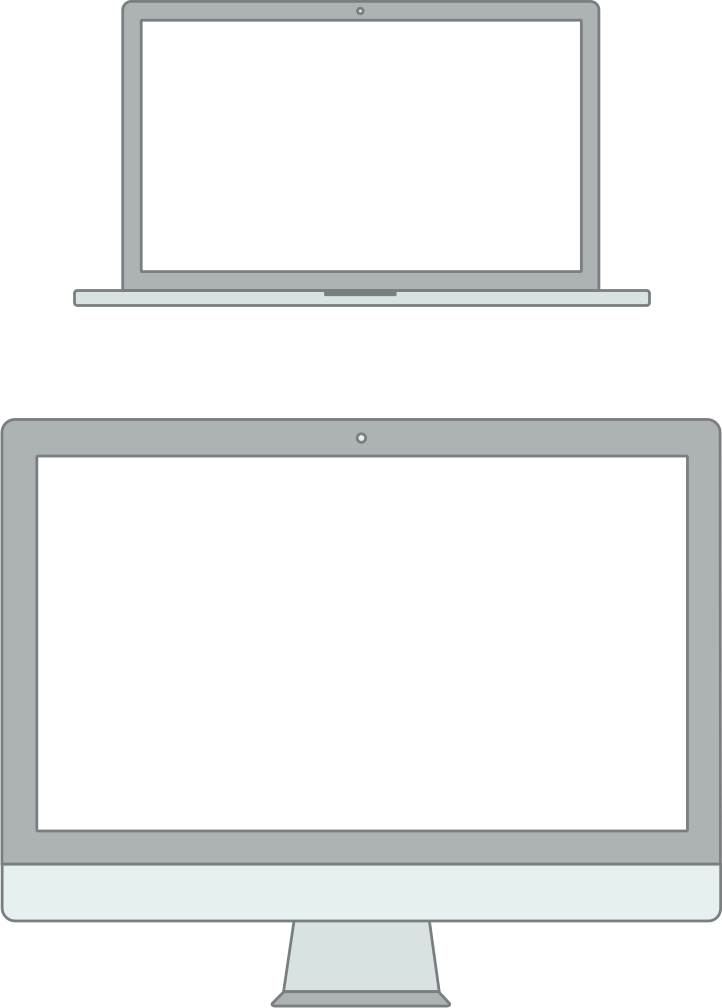
Windows and pointers.
Multi-tasking, multiple windows.
Exposes and leverages the file system.
Many specialized tools in concert.
Underneath, it’s the 1970s, UNIX, and the command-line.
Cabinets, drawers, and files model of where things are stored
Where technical computing lives

This toolset is by now really good!
Free! Open! Powerful!
Friendly communities! Lots of information! Many resources!
But: grounded in a UI paradigm that is increasingly far away from the everyday use of computing devices
So why do we use this stuff?
“Office” vs “Engineering” approaches
What is “real” in your project?
What is the final output?
How is it produced?
How are changes managed?
Different Answers
Office model
- Formatted documents are real.
- Intermediate outputs are cut and pasted into documents.
- Changes are tracked inside files.
- Final output is often in the same format you’ve been working in, e.g. a Word file, or a PDF.
Engineering model
- Plain-text files are real.
- Intermediate outputs are produced via code, often inside documents.
- Changes are tracked outside files, at the level of a project.
- Final outputs are assembled programmatically and converted to some desired format.
Different strengths and weaknesses
Office model
Everyone knows Word, Excel, or Google Docs.
“Track changes” is powerful and easy.
Hm, I can’t remember how I made this figure
Where did this table of results come from?
Paper_edits_FINAL_kh-1.docx
Engineering model
Plain text is highly portable.
Push button, recreate analysis.
JFC Why can’t I do this simple thing?
Object of type 'closure' is not subsettable
Each approach generates solutions to its own problems
The File System
The traditional analog

The problem is, you probably have never have actually used one of these!
The file cabinet!

The file cabinet!

Index cards

Index cards

Automating information processing

Automating information processing

Automating information processing

Hollerith machines

Hollerith Machines

Hollerith machines

Hollerith machines

Hollerith Operators

Hollerith Operators

IBM punch cards

IBM punch cards

Big Iron

Storage

Storage

Input/Output
A late-model teletype (TTY) machine

Input/Output
The DEC VT-100 Terminal

Input/Output
Back to the file system
File system hierarchy

Stepping back
Your computer stores files and does stuff, or “runs commands”
Files are stored in a large hierarchy of folders
The Finder or Window Manager or File Manager is a visual metaphor for representing this hierarchy of files and for running commands on them. But you can also do these things via text-based commands delivered from a prompt, console, or “command line”.
Software like RStudio has a lot of these “old school” computing elements
Getting to know R and RStudio
We want to draw graphs reproducibly


Abstraction in software
Less
- Easy things are awkward
- Hard things are straightforward
- Really hard things are possible
Abstraction in software
Less
Easy things are awkward
Hard things are straightforward
Really hard things are possible
More
Easy things are trivial
Hard things are awkward
Really hard things are impossible
Compare
- D3
- Grid
- ggplot
- Stata
- Excel
The RStudio IDE
An IDE for R
An IDE for Meals
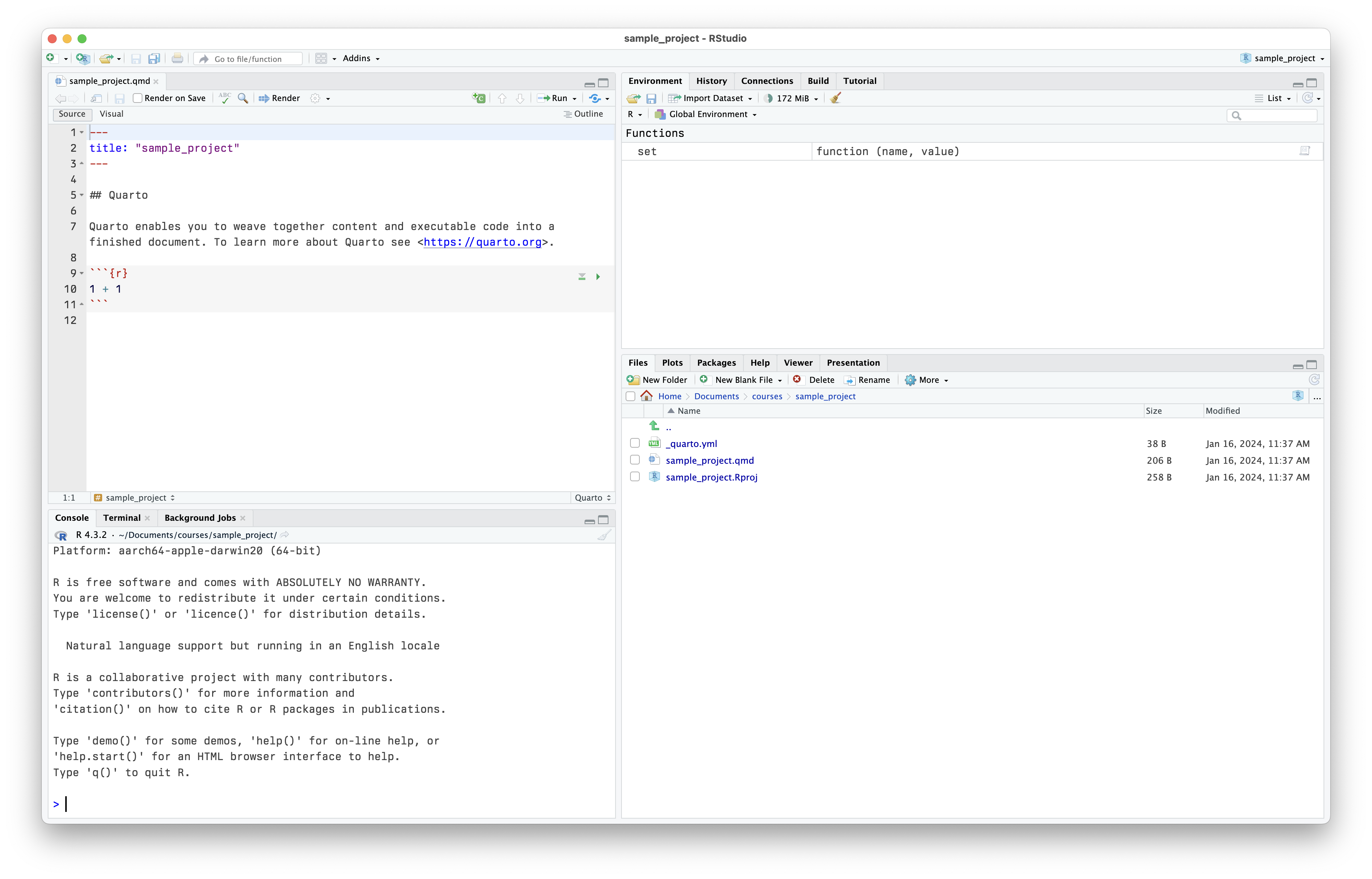
RStudio at startup
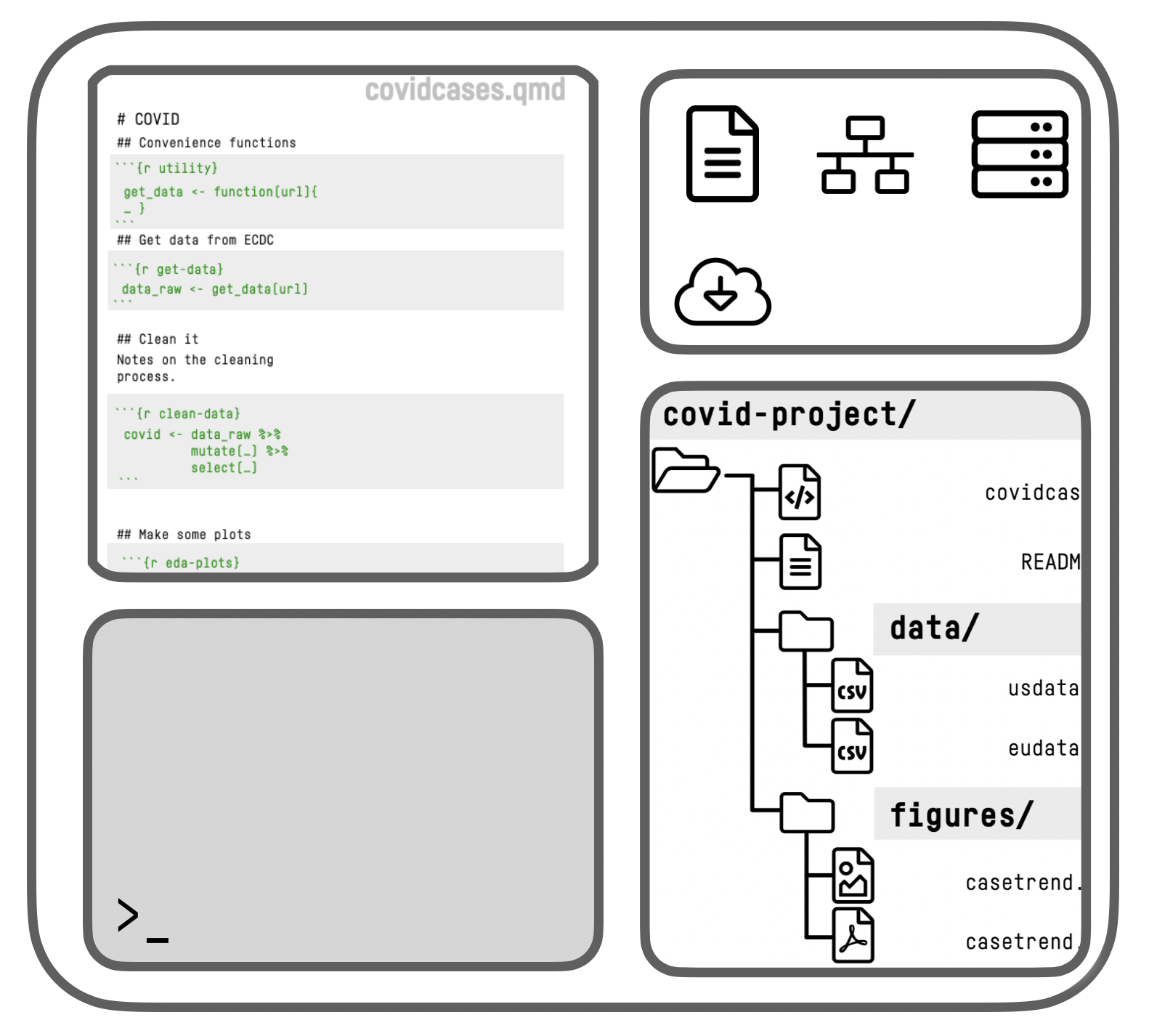
RStudio schematic overview
RStudio schematic overview
Think in terms of Data + Transformations, written out as code, rather than a series of point-and-click steps
Our starting data + our code is what’s “real” in our projects, not the final output or any intermediate objects

RStudio at startup

RStudio at startup
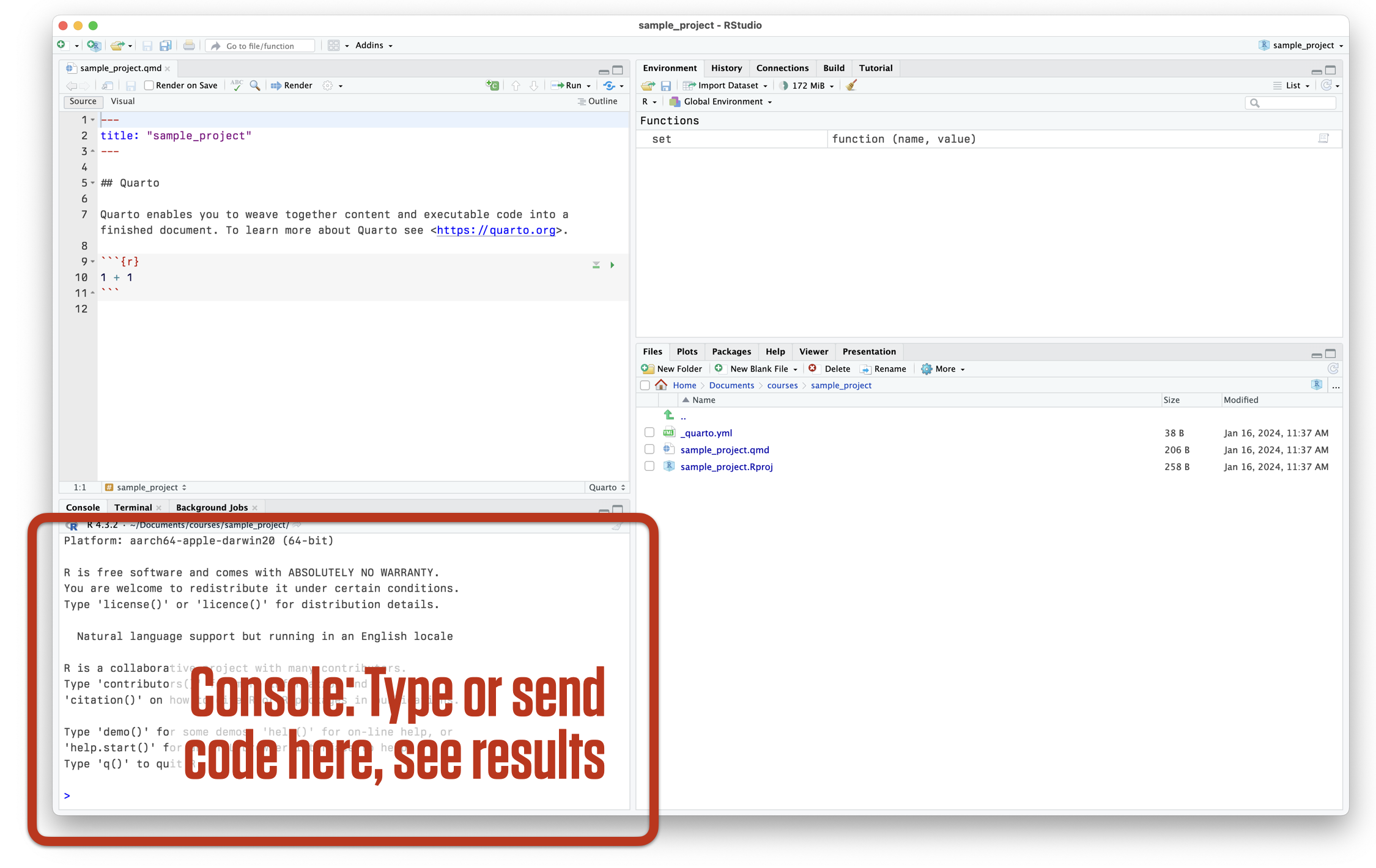
RStudio at startup
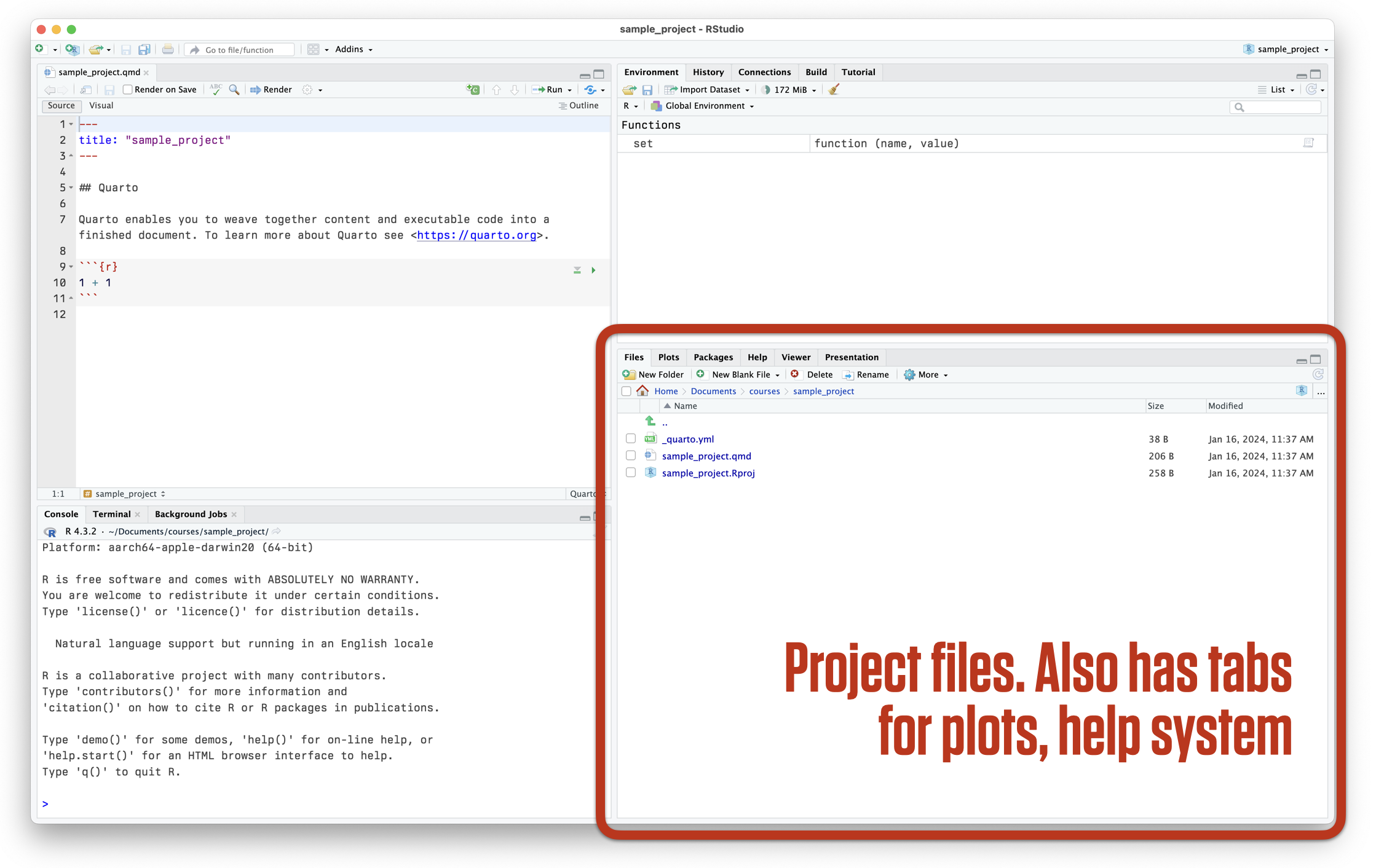
RStudio at startup
RStudio at startup
Use RMarkdown to produce and reproduce work
Where we want to end up

PDF out
Where we want to end up

HTML out
Where we want to end up

Word out
How to get there?

- We could write an R script with some notes inside, using it to create some figures and tables, paste them into our document.
- This will work, but we can do better.
We can make this …
… by writing this

The code gets replaced by its output



Markdown document

Markdown document annotated

- This approach has its limitations, but it’s very useful and has many benefits.
Basic markdown summary
| Desired style | Use the following Markdown annotation |
|---|---|
| Heading 1 | # Heading 1 |
| Heading 2 | ## Heading 2 |
| Heading 3 | ### Heading 3 (Actual heading styles will vary.) |
| Paragraph | Just start typing |
| Bold | **Bold** |
| Italic | *Italic* |
| Images | [Alternate text for image](path/image.jpg) |
| Hyperlinks | [Link text](https://www.visualizingsociety.com/) |
| Unordered Lists | |
| - First | - First |
| - Second. | - Second |
| - Third | - Third |
| Ordered Lists | |
| 1. First | 1. First |
| 2. Second. | 2. Second |
| 3. Third | 3. Third |
| Footnote.¹ | Footnote[^notelabel] |
| ¹The note’s content. | [^notelabel] The note's content. |
The right frame of mind
- This is like learning how to drive a car, or how to cook in a kitchen … or learning to speak a language.
- After some orientation to what’s where, you will learn best by doing.
- Software is a pain, but you won’t crash the car or burn your house down.
TYPE OUT
YOUR CODE
BY HAND

Samuel Beckett
GETTING ORIENTED
Loading the tidyverse libraries
- The tidyverse has several components.
- We’ll return to this message about Conflicts later.
- Again, the code and messages you see here is actual R output, produced at the same time as the slide.
Tidyverse components
library(tidyverse)Loading tidyverse: ggplot2Loading tidyverse: tibbleLoading tidyverse: tidyrLoading tidyverse: readrLoading tidyverse: purrrLoading tidyverse: dplyr
- Call the package and …
<|Draw graphs<|Nicer data tables<|Tidy your data<|Get data into R<|Fancy Iteration<|Action verbs for tables
What R looks like
Code you can type and run:
Output:
This is equivalent to running the code above, typing my_numbers at the console, and hitting enter.
What R looks like
By convention, code output in documents is prefixed by ##
Also by convention, outputting vectors, etc, gets a counter keeping track of the number of elements. For example,
Some things to know about R
0. It’s a calculator
0. It’s a calculator
Boolean and Logical operators
Logical equality and inequality (yielding a TRUE or FALSE result) is done with == and !=. Other logical operators include <, >, <=, >=, and ! for negation.
1. Everything in R has a name
Some names are forbidden
Or it’s a really bad idea to try to use them
2. Everything is an object
There are a few built-in objects:
[1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" "m" "n" "o" "p" "q" "r" "s"
[20] "t" "u" "v" "w" "x" "y" "z"3. You can create objects
In fact, this is mostly what we will be doing.
Objects are created by assigning a thing to a name:
The c() function combines or concatenates things
The assignment operator
- The assignment operator performs the action of creating objects
- Use a keyboard shortcut to write it:
- Press
optionand-on a Mac - Press
altand-on Windows
Assignment with =
- You can use
=as well as<-for assignment.
- On the other hand,
=has a different meaning when used in functions. - I’m going to use
<-for assignment throughout. - Be consistent either way.
4. You do things with functions
4. You do things with functions
- Functions can be identified by the parentheses after their names.
What functions usually do
- They take inputs to arguments
- They perform actions
- They produce, or return, outputs
mean(x = my_numbers)
What functions usually do
- They take inputs to arguments
- They perform actions
- They produce, or return, outputs
mean(x = my_numbers)
[1] 6.25
What functions usually do
What functions usually do
If you don’t name the arguments, R assumes you are providing them in the order the function expects.
What functions usually do
What arguments? Which order? Read the function’s help page
- How to read an R help page?
What functions usually do
- Arguments often tell the function what to do in specific circumstances
[1] NA[1] 32.44444Or select from one of several options
What functions usually do
There are all kinds of functions. They return different things.
What functions usually do
You can assign the output of a function to a name, which turns it into an object. (Otherwise it’ll send its output to the console.)
What functions usually do
Objects hang around in your work environment until they are overwritten by you, or are deleted.
Functions can be nested
Nested functions are evaluated from the inside out.
Use the pipe operator: |>
Instead of deeply nesting functions in parentheses, we can use the pipe operator:
Read this operator as “and then”
Use the pipe operator: |>
Better, vertical space is free in R:
Pipelines make code more readable
Not great, Bob:
Notice how the first thing you read is the last operation performed.
Pipelines make code more readable
We can use vertical space and indents, but it’s really not much better:
Pipelines make code more readable
Much nicer:
eggs |>
get_from_fridge() |>
crack_eggs(into = "bowl") |>
whisk(len = 40) |>
pour_in_pan(temp = "med-high") |>
stir() |>
serve()- We’ll still use nested parentheses quite a bit, often in the context of a function working inside a pipeline. But it’s good not to have too many levels of nesting.
The other pipe: %>%
The Base R pipe operator, |> is a relatively recent addition to R.
Piping operations were originally introduced in a package called called magrittr, where it took the form %>%
The other pipe: %>%
The Base R pipe operator, |> is a relatively recent addition to R.
Piping operations were originally introduced in a package called called magrittr, where it took the form %>%
It’s been so successful, a version of it has been incorporated into Base R. For our puposes, they’re the same.
Functions are bundled into packages
Packages are loaded into your working environment using the library() function:
# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsFunctions are bundled into packages
You need only install a package once (and occasionally update it):
Functions are bundled into packages
But you must load the package in each R session before you can access its contents:
## To load a package, usually at the start of your RMarkdown document or script file
library(palmerpenguins)
penguins# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
7 Adelie Torgersen 38.9 17.8 181 3625
8 Adelie Torgersen 39.2 19.6 195 4675
9 Adelie Torgersen 34.1 18.1 193 3475
10 Adelie Torgersen 42 20.2 190 4250
# ℹ 334 more rows
# ℹ 2 more variables: sex <fct>, year <int>Let’s Go!
Like before
What we did
Load the packages we need: tidyverse and gapminder
What we did
New object named p gets the output of the ggplot() function, given these arguments
Notice how one of the arguments, mapping, is itself taking the output of a function named aes()
What we did
Show me the output of the p object and the geom_point() function.
The + here acts just like the |> pipe, but for ggplot functions only. (This is an accident of history.)
And what is R doing?
R objects are just lists of stuff to use or things to do
Objects are like Bento Boxes
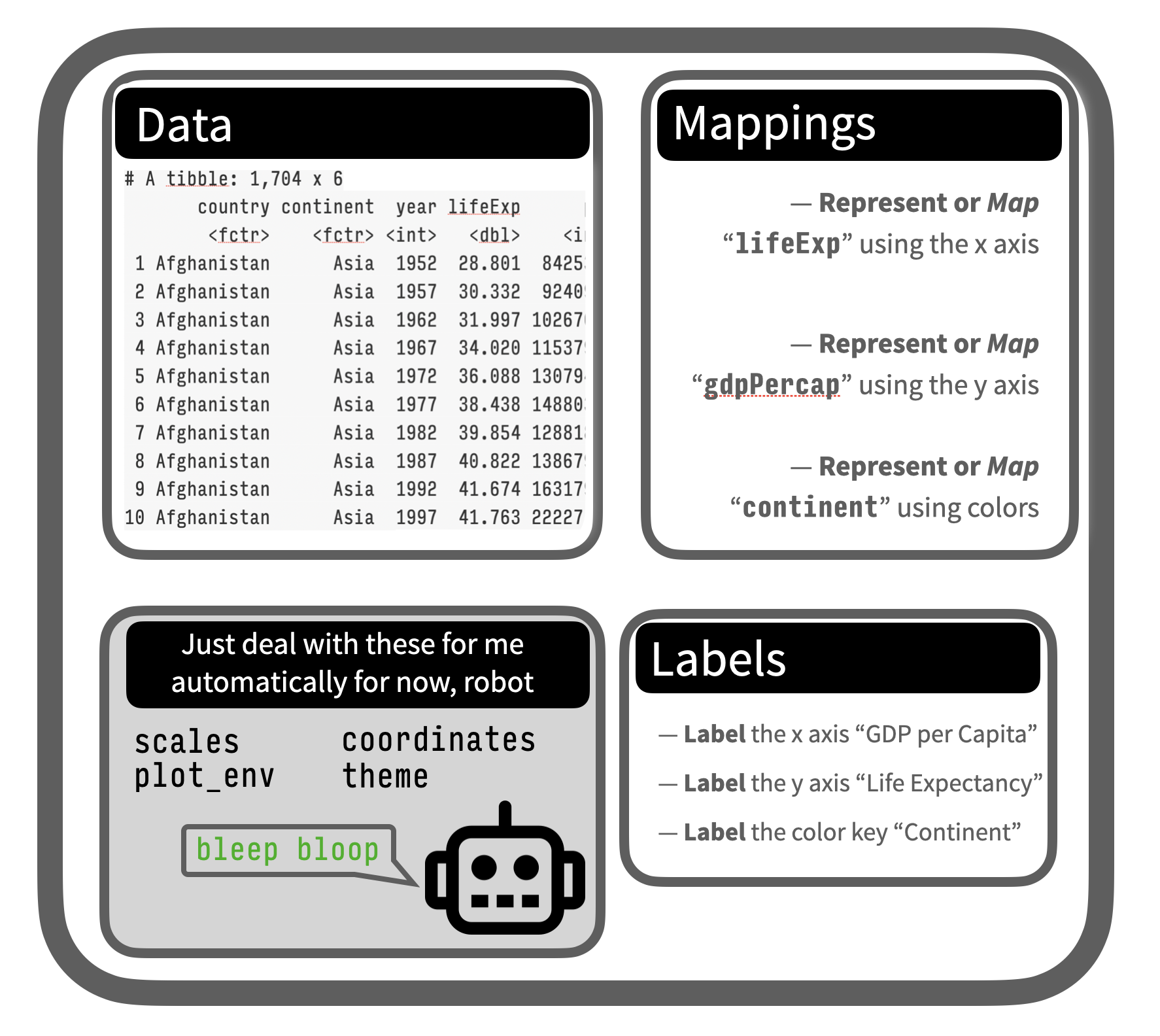
The p object
Peek in with the object inspector
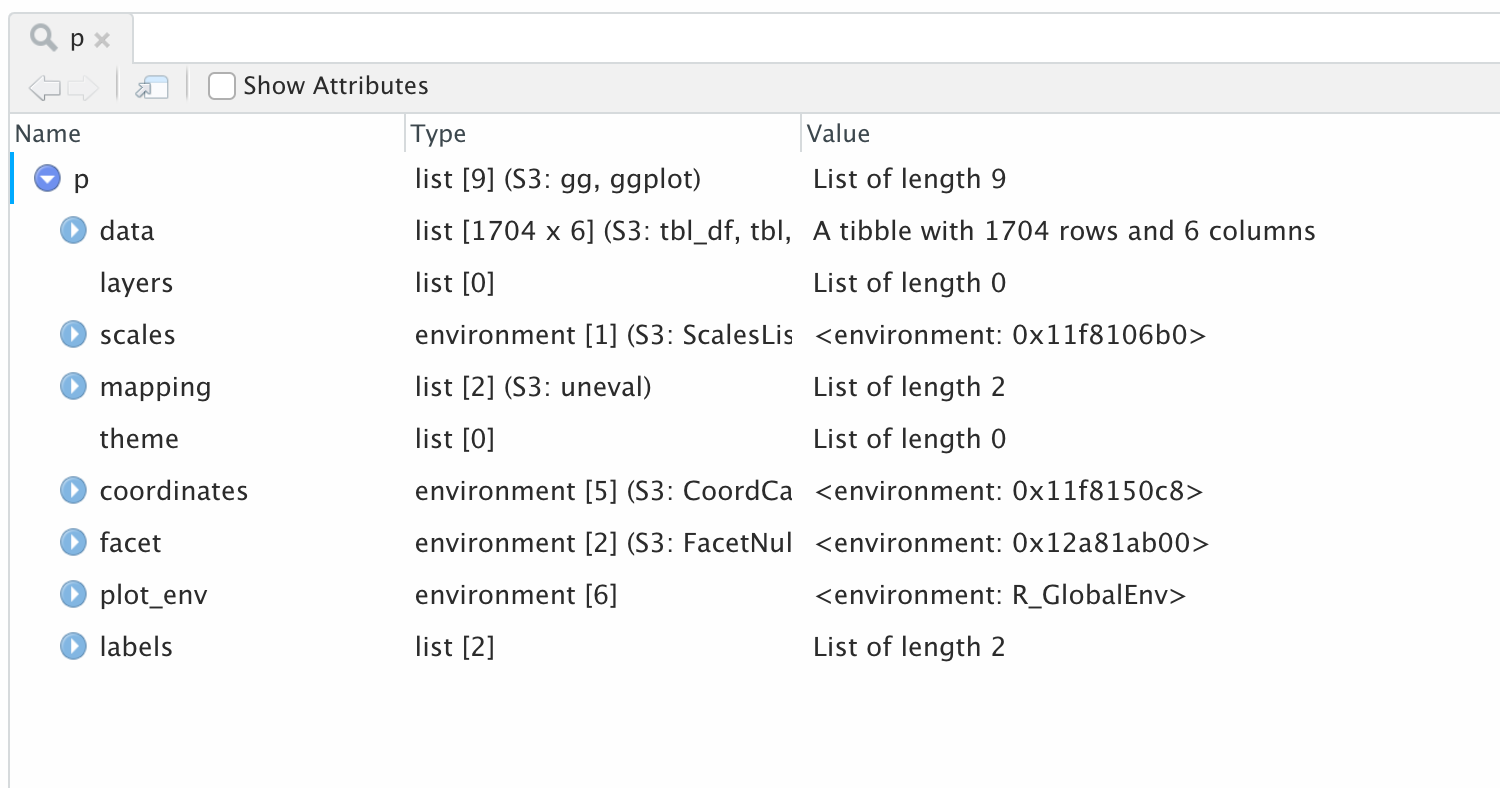
Peek in with the object inspector
Core concepts: mappings + geoms
The core idea, which we’ll focus on more formally next week, is that we have data, arranged in columns, that we want to represent visually on some sort of plot.
That means we need a mapping — a link, a connection, a representation — between things in our table and stuff we can draw. That is what the mapping argument is for.
And we need a geom — a kind of plot, a particular sort of graph — that we draw with that.
Practical examples
Let’s try some live examples …How might we improve or extend this graph based on the data we have? Or how might we look at it differently?

```
