library(here) # manage file paths
library(socviz) # data and some useful functions
library(tidyverse) # your friend and mine06 — Extend your Vocabulary
February 14, 2024
Extend your
ggplot
vocabulary
Load our libraries
Tidyverse components
library(tidyverse)Loading tidyverse: ggplot2Loading tidyverse: tibbleLoading tidyverse: tidyrLoading tidyverse: readrLoading tidyverse: purrrLoading tidyverse: dplyr
- Load the package and …
<|Draw graphs<|Nicer data tables<|Tidy your data<|Get data into R<|Fancy Iteration<|Action verbs for tables
Other tidyverse components
forcatshavenlubridatereadxlstringrreprex
<|Deal with factors<|Import Stata, SPSS, etc<|Dates, Durations, Times<|Import from spreadsheets<|Strings and Regular Expressions<|Make reproducible examples
Not all of these are attached when we do library(tidyverse)
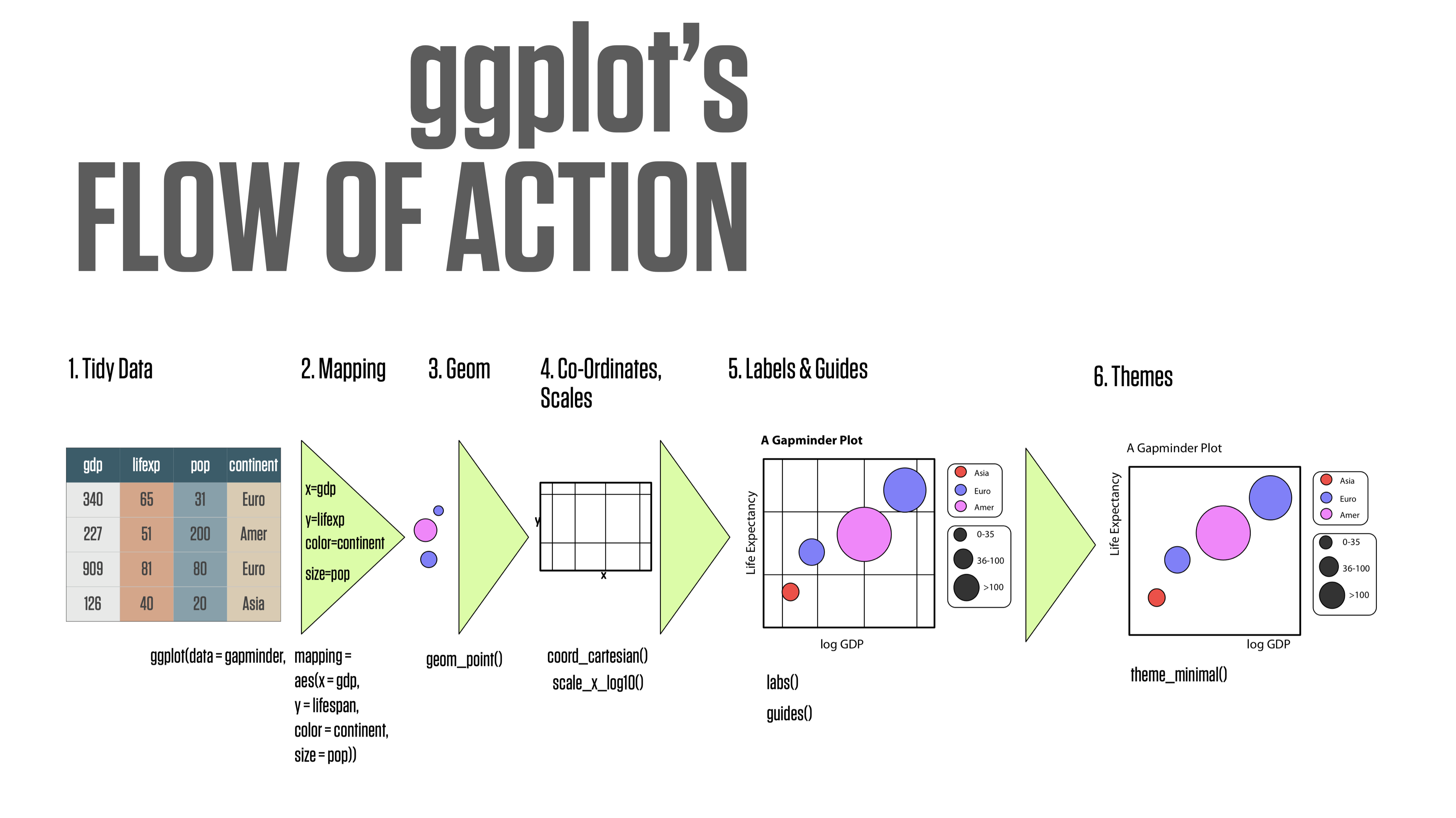
ggplot’s flow of action
Thinking in terms of layers
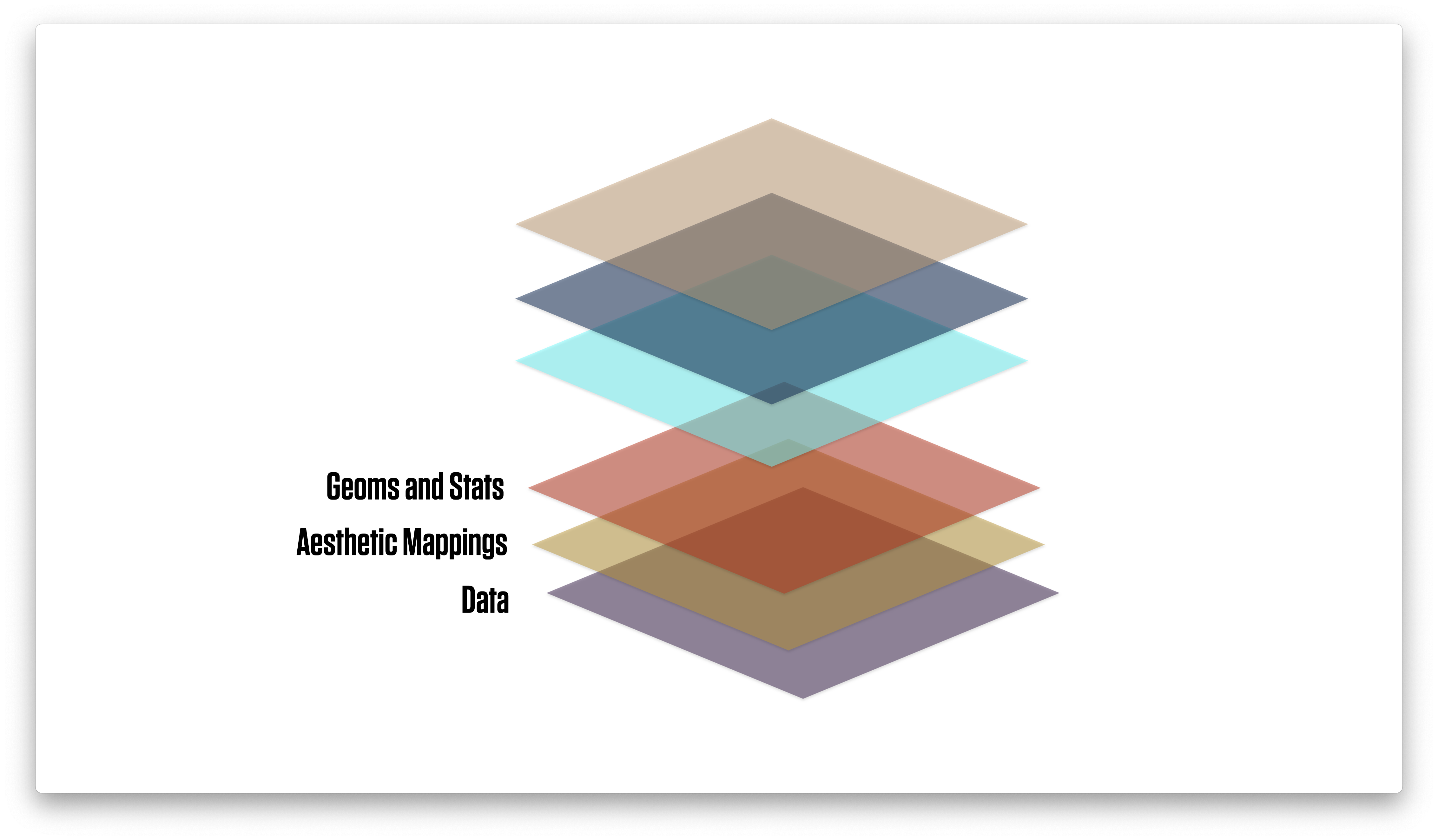
Thinking in terms of layers
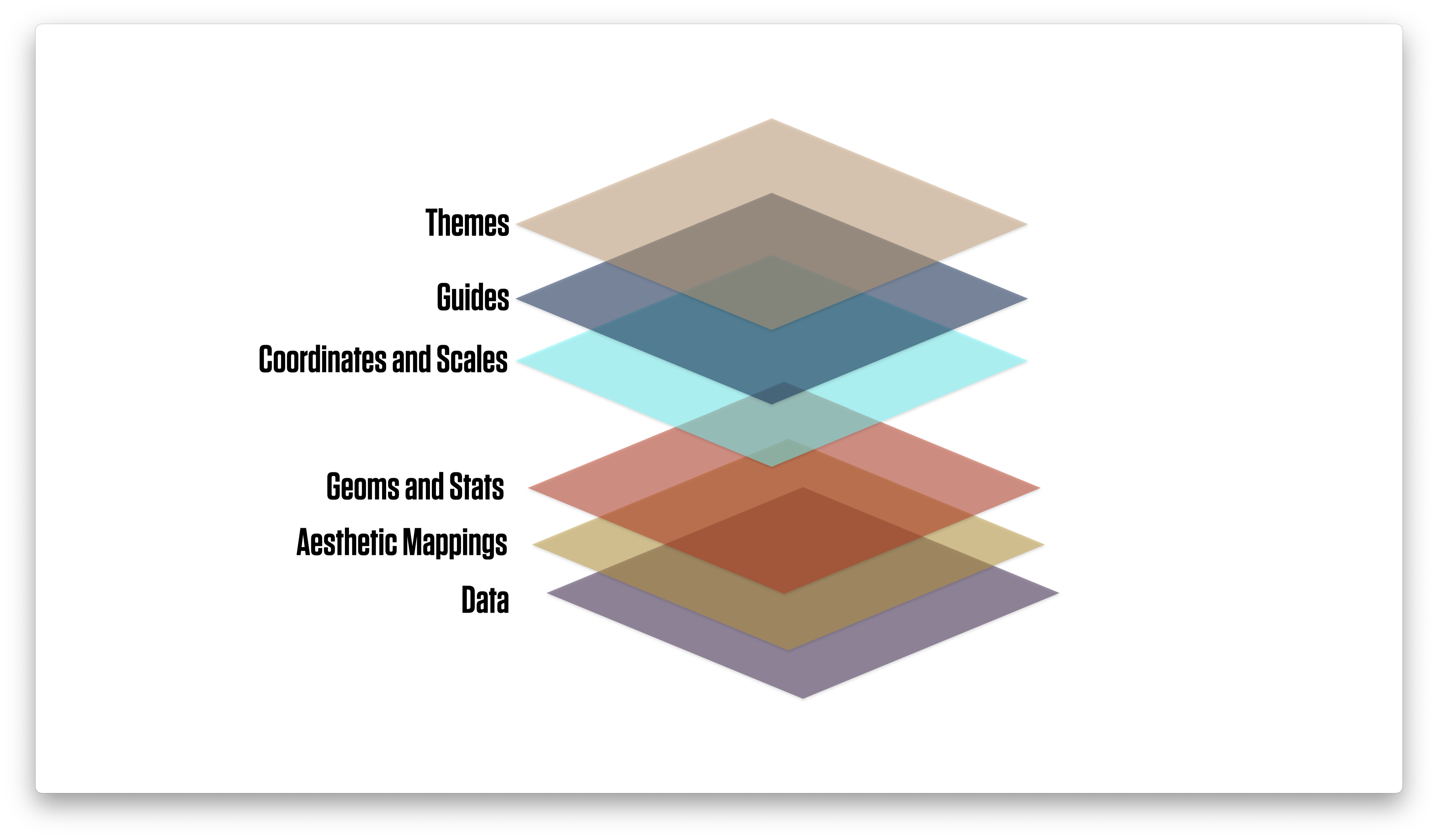
Thinking in terms of layers
Feeding data
to ggplot
Transform and summarize first.
Then send your clean tables to ggplot.
Extend your
ggplot vocabulary
We’ll move forward in three ways
Learn more geoms
geom_point(),geom_line(),geom_col(),geom_histogram(),geom_density(),geom_jitter(),geom_boxplot(),geom_pointrange(),…
We’ll move forward in three ways
Learn more geoms
geom_point(),geom_line(),geom_col(),geom_histogram(),geom_density(),geom_jitter(),geom_boxplot(),geom_pointrange(),…
Learn more about scales, guides, and themes
- Functions that control the details of representing data and styling our plots.
We’ll move forward in three ways
Learn more geoms
geom_point(),geom_line(),geom_col(),geom_histogram(),geom_density(),geom_jitter(),geom_boxplot(),geom_pointrange(),…
Learn more about scales, guides, and themes
- Functions that control the details of representing data and styling our plots.
Learn more about extensions to ggplot
- Packages that enhance
ggplot’s capabilities, usually by adding support for new kinds of plot (i.e., new geoms), or new functionality (e.g., thescalespackage).
Example and extension:
Organ Donation data
organdata is in the socviz package
# A tibble: 238 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Australia NA NA 17065 0.220 16774 16591 1300 1224
2 Australia 1991-01-01 12.1 17284 0.223 17171 16774 1379 1300
3 Australia 1992-01-01 12.4 17495 0.226 17914 17171 1455 1379
4 Australia 1993-01-01 12.5 17667 0.228 18883 17914 1540 1455
5 Australia 1994-01-01 10.2 17855 0.231 19849 18883 1626 1540
6 Australia 1995-01-01 10.2 18072 0.233 21079 19849 1737 1626
7 Australia 1996-01-01 10.6 18311 0.237 21923 21079 1846 1737
8 Australia 1997-01-01 10.3 18518 0.239 22961 21923 1948 1846
9 Australia 1998-01-01 10.5 18711 0.242 24148 22961 2077 1948
10 Australia 1999-01-01 8.67 18926 0.244 25445 24148 2231 2077
# ℹ 228 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>First look

First look

First look

First look

First look

First look

Showing continuous measures by category
Boxplots: geom_boxplot()

Put categories on the y-axis!

Reorder y by the mean of x

(Reorder y by any statistic you like)

geom_boxplot() can color and fill

These strategies are quite general

geom-jitter() for overplotting

Adjust with a position argument

Using across() and where()
by_country <- organdata |>
group_by(consent_law, country) |>
summarize(across(where(is.numeric),
list(mean = \(x) mean(x, na.rm = TRUE),
sd = \(x) sd(x, na.rm = TRUE))),
.groups = "drop")
head(by_country) # A tibble: 6 × 28
consent_law country donors_mean donors_sd pop_mean pop_sd pop_dens_mean
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Informed Australia 10.6 1.14 18318. 831. 0.237
2 Informed Canada 14.0 0.751 29608. 1193. 0.297
3 Informed Denmark 13.1 1.47 5257. 80.6 12.2
4 Informed Germany 13.0 0.611 80255. 5158. 22.5
5 Informed Ireland 19.8 2.48 3674. 132. 5.23
6 Informed Netherlands 13.7 1.55 15548. 373. 37.4
# ℹ 21 more variables: pop_dens_sd <dbl>, gdp_mean <dbl>, gdp_sd <dbl>,
# gdp_lag_mean <dbl>, gdp_lag_sd <dbl>, health_mean <dbl>, health_sd <dbl>,
# health_lag_mean <dbl>, health_lag_sd <dbl>, pubhealth_mean <dbl>,
# pubhealth_sd <dbl>, roads_mean <dbl>, roads_sd <dbl>, cerebvas_mean <dbl>,
# cerebvas_sd <dbl>, assault_mean <dbl>, assault_sd <dbl>,
# external_mean <dbl>, external_sd <dbl>, txp_pop_mean <dbl>,
# txp_pop_sd <dbl>Plot our summary data
What about faceting it instead?
The problem is that countries can only be in one Consent Law category.
What about faceting it instead?
Restricting to one column doesn’t fix it.
Allow the y-scale to vary
Normally the point of a facet is to preserve comparability between panels by not allowing the scales to vary. But for categorical measures it can be useful to allow this.
Again, these methods are general
by_country |>
ggplot(mapping =
aes(x = donors_mean,
y = reorder(country, donors_mean),
color = consent_law)) +
geom_pointrange(mapping =
aes(xmin = donors_mean - donors_sd,
xmax = donors_mean + donors_sd)) +
guides(color = "none") +
facet_wrap(~ consent_law,
ncol = 1,
scales = "free_y") +
labs(x = "Donor Procurement Rate",
y = NULL,
color = "Consent Law")
Your turn
Load this data
# A tibble: 4,343 × 9
title year runtime maturity_rating genre box_office rating_imdb metascore
<chr> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 102 Dal… 2000 100 G Fami… 67 4.8 35
2 28 Days 2000 103 PG-13 Come… 37.2 6.1 46
3 3 Strik… 2000 82 R Come… 9.8 4.6 11
4 A Shot … 2000 114 R Sport 0.1 6.2 66
5 About A… 2000 97 R Come… 0.2 5.8 64
6 All the… 2000 116 PG-13 West… 15.5 5.8 55
7 Almost … 2000 122 R Come… 32.5 7.9 90
8 America… 2000 102 R Horr… 15.1 7.6 64
9 An Ever… 2000 103 R Come… 0.1 6.2 56
10 Autumn … 2000 103 PG-13 Roma… 37.8 5.6 24
# ℹ 4,333 more rows
# ℹ 1 more variable: awards <dbl>Overview
English-language movies produced in the US; at least 80 minutes long and no longer than 3.5 hours; received at least 500 votes on the Internet Movie Database; MPAA rating between G and R; made at least $100,000 domestically
Overview
- year: The calendar year of the film’s release.
- runtime: The length of the movie in minutes.
- maturity_rating: The movie’s MPA maturity rating (G, PG, PG-13, or R).
- genre: The genre of the film (one only).
- box_office: Gross domestic (US only) box office returns for the movie in millions of US dollars. Not adjusted for inflation.
- rating_imdb: This is average score (between 1 and 10) for a movie provided by IMDB users.
- metascore: The movie’s metascore rating from metacritic. The metascore is a curated weighted average of reviewer scores from a variety of sources.
- awards: The number of Oscar awards that this movie received.
What can we learn from visualizing this data?
Plot text directly
geom_text() for basic labels
It’s not very flexible
There are tricks, but they’re limited
We’ll use ggrepel instead
The ggrepel package provides geom_text_repel() and geom_label_repel()
Example: U.S. Historic
Presidential Elections
elections_historic is in socviz
# A tibble: 49 × 19
election year winner win_party ec_pct popular_pct popular_margin votes
<int> <int> <chr> <chr> <dbl> <dbl> <dbl> <int>
1 10 1824 John Quinc… D.-R. 0.322 0.309 -0.104 1.13e5
2 11 1828 Andrew Jac… Dem. 0.682 0.559 0.122 6.43e5
3 12 1832 Andrew Jac… Dem. 0.766 0.547 0.178 7.03e5
4 13 1836 Martin Van… Dem. 0.578 0.508 0.142 7.63e5
5 14 1840 William He… Whig 0.796 0.529 0.0605 1.28e6
6 15 1844 James Polk Dem. 0.618 0.495 0.0145 1.34e6
7 16 1848 Zachary Ta… Whig 0.562 0.473 0.0479 1.36e6
8 17 1852 Franklin P… Dem. 0.858 0.508 0.0695 1.61e6
9 18 1856 James Buch… Dem. 0.588 0.453 0.122 1.84e6
10 19 1860 Abraham Li… Rep. 0.594 0.396 0.101 1.86e6
# ℹ 39 more rows
# ℹ 11 more variables: margin <int>, runner_up <chr>, ru_part <chr>,
# turnout_pct <dbl>, winner_lname <chr>, winner_label <chr>, ru_lname <chr>,
# ru_label <chr>, two_term <lgl>, ec_votes <dbl>, ec_denom <dbl>We’ll draw a plot like this
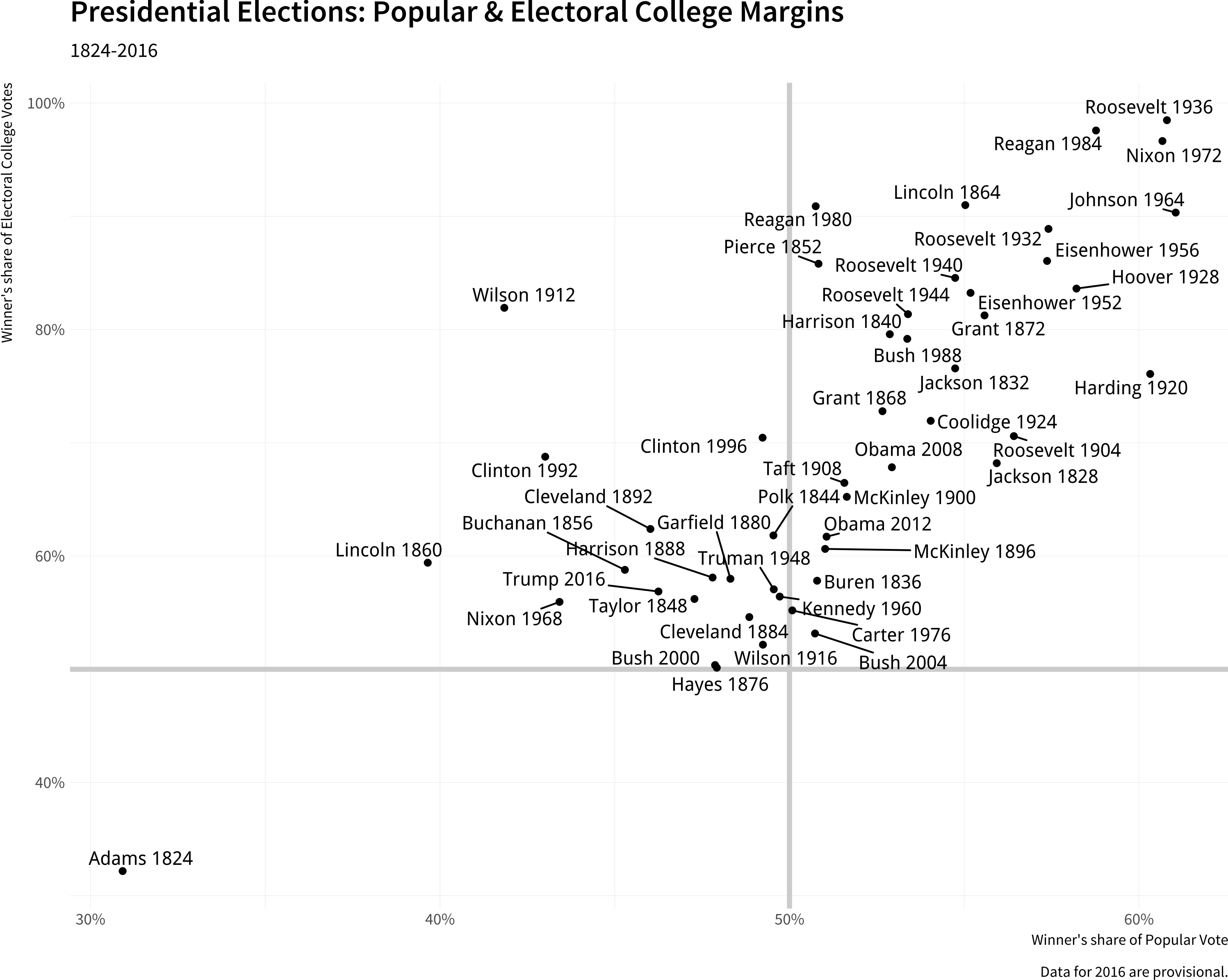
Presidential elections
Keep things neat
## The packages we'll use in addition to ggplot
library(ggrepel)
library(scales)
p_title <- "Presidential Elections: Popular & Electoral College Margins"
p_subtitle <- "1824-2016"
p_caption <- "Data for 2016 are provisional."
x_label <- "Winner's share of Popular Vote"
y_label <- "Winner's share of Electoral College Votes"Base Layer, Lines, Points
Add the labels
This looks terrible here because geom_text_repel() uses the dimensions of the available graphics device to iteratively figure out the labels. Let’s allow it to draw on the whole slide.
Labeling is with respect to the plot size

Adjust the Scales
p <- ggplot(data = elections_historic,
mapping = aes(x = popular_pct,
y = ec_pct,
label = winner_label))
p_out <- p + geom_hline(yintercept = 0.5,
linewidth = 1.4,
color = "gray80") +
geom_vline(xintercept = 0.5,
linewidth = 1.4,
color = "gray80") +
geom_point() +
geom_text_repel() +
scale_x_continuous(labels = label_percent()) +
scale_y_continuous(labels = label_percent()) 
Add the labels
p <- ggplot(data = elections_historic,
mapping = aes(x = popular_pct,
y = ec_pct,
label = winner_label))
p_out <- p + geom_hline(yintercept = 0.5,
linewidth = 1.4,
color = "gray80") +
geom_vline(xintercept = 0.5,
linewidth = 1.4,
color = "gray80") +
geom_point() +
geom_text_repel(mapping = aes(family = "Tenso Slide")) +
scale_x_continuous(labels = label_percent()) +
scale_y_continuous(labels = label_percent()) +
labs(x = x_label, y = y_label,
title = p_title,
subtitle = p_subtitle,
caption = p_caption) 
Labeling points
of interest
Option 1: On the fly in ggplot
Option 1: On the fly inside ggplot
Stuffing everything into the subset() call might get messy
Option 2: Use dplyr first
# A tibble: 6 × 28
consent_law country donors_mean donors_sd pop_mean pop_sd pop_dens_mean
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Informed Ireland 19.8 2.48 3674. 132. 5.23
2 Informed United States 20.0 1.33 269330. 12545. 2.80
3 Presumed Belgium 21.9 1.94 10153. 109. 30.7
4 Presumed Norway 15.4 1.11 4386. 97.3 1.35
5 Presumed Spain 28.1 4.96 39666. 951. 7.84
6 Presumed Switzerland 14.2 1.71 7037. 170. 17.0
# ℹ 21 more variables: pop_dens_sd <dbl>, gdp_mean <dbl>, gdp_sd <dbl>,
# gdp_lag_mean <dbl>, gdp_lag_sd <dbl>, health_mean <dbl>, health_sd <dbl>,
# health_lag_mean <dbl>, health_lag_sd <dbl>, pubhealth_mean <dbl>,
# pubhealth_sd <dbl>, roads_mean <dbl>, roads_sd <dbl>, cerebvas_mean <dbl>,
# cerebvas_sd <dbl>, assault_mean <dbl>, assault_sd <dbl>,
# external_mean <dbl>, external_sd <dbl>, txp_pop_mean <dbl>,
# txp_pop_sd <dbl>Option 2: Use dplyr first
This makes things neater. A geom can be fully “autonomous”. Each one can have its own mapping call and its own data source. This can be very useful when building up plots overlaying several sources or subsets of data.
Write and draw
inside the plot area
annotate() can imitate geoms
annotate() can imitate geoms
Scales, Guides, and Themes
Every mapped variable has a scale
- Aesthetic mappings link quantities or categories in your data to things you can see on the graph. Thus, they have a scale associated with that representation.
- Scale functions manage this relationship. Remember: not just
xandybut alsocolor,fill,shape,size, andalphaare scales. - If it can represent your data, it has a scale, and a scale function to manage it.
- This means you control things like color schemes for data mappings through scale functions
- Because those colors are representing features of your data.
Naming conventions for scale functions
- In general, scale functions are named like this:
scale_<MAPPING>_<KIND>()
Naming conventions
- In general, scale functions are named like this:
scale_<MAPPING>_<KIND>()
- We already know there are a lot of mappings
- x, y, color, size, shape, and so on.
Naming conventions
- In general, scale functions are named like this:
scale_<MAPPING>_<KIND>()
We already know there are a lot of mappings
x, y, color, size, shape, and so on.
And there are many kinds of scale as well.
discrete, continuous, log10, date, binned, and many others.
So there’s a whole zoo of scale functions.
The naming convention helps us keep track.
Naming conventions
scale_<MAPPING>_<KIND>()
- scale_x_continuous()
- scale_y_continous()
- scale_x_discrete()
- scale_y_discrete()
- scale_x_log10()
- scale_x_sqrt()
Naming conventions
scale_<MAPPING>_<KIND>()
- scale_x_continuous()
- scale_y_continous()
- scale_x_discrete()
- scale_y_discrete()
- scale_x_log10()
- scale_x_sqrt()
- scale_color_discrete()
- scale_color_gradient()
- scale_color_gradient2()
- scale_color_brewer()
- scale_fill_discrete()
- scale_fill_gradient()
- scale_fill_gradient2()
- scale_fill_brewer()
Scale functions in practice
- Scale functions take arguments appropriate to their mapping and kind
More usefully …
The guides() function
- Control overall properties of the guide labels.
- Common use: turning it off.
- We’ll see more advanced uses later.
The theme() function
theme() styles parts of your plot that are not directly representing your data. Often the first thing people want to adjust; but logically it’s the last thing.
## Using the "classic" ggplot theme here
organdata |>
ggplot(mapping = aes(x = roads,
y = donors,
color = consent_law)) +
geom_point() +
labs(title = "By Consent Law",
x = "Road Deaths",
y = "Donor Procurement",
color = "Legal Regime:") +
theme(legend.position = "bottom",
plot.title = element_text(color = "darkred",
face = "bold"))
Sidenote: Smoothers

A trend
Sidenote: Smoothers
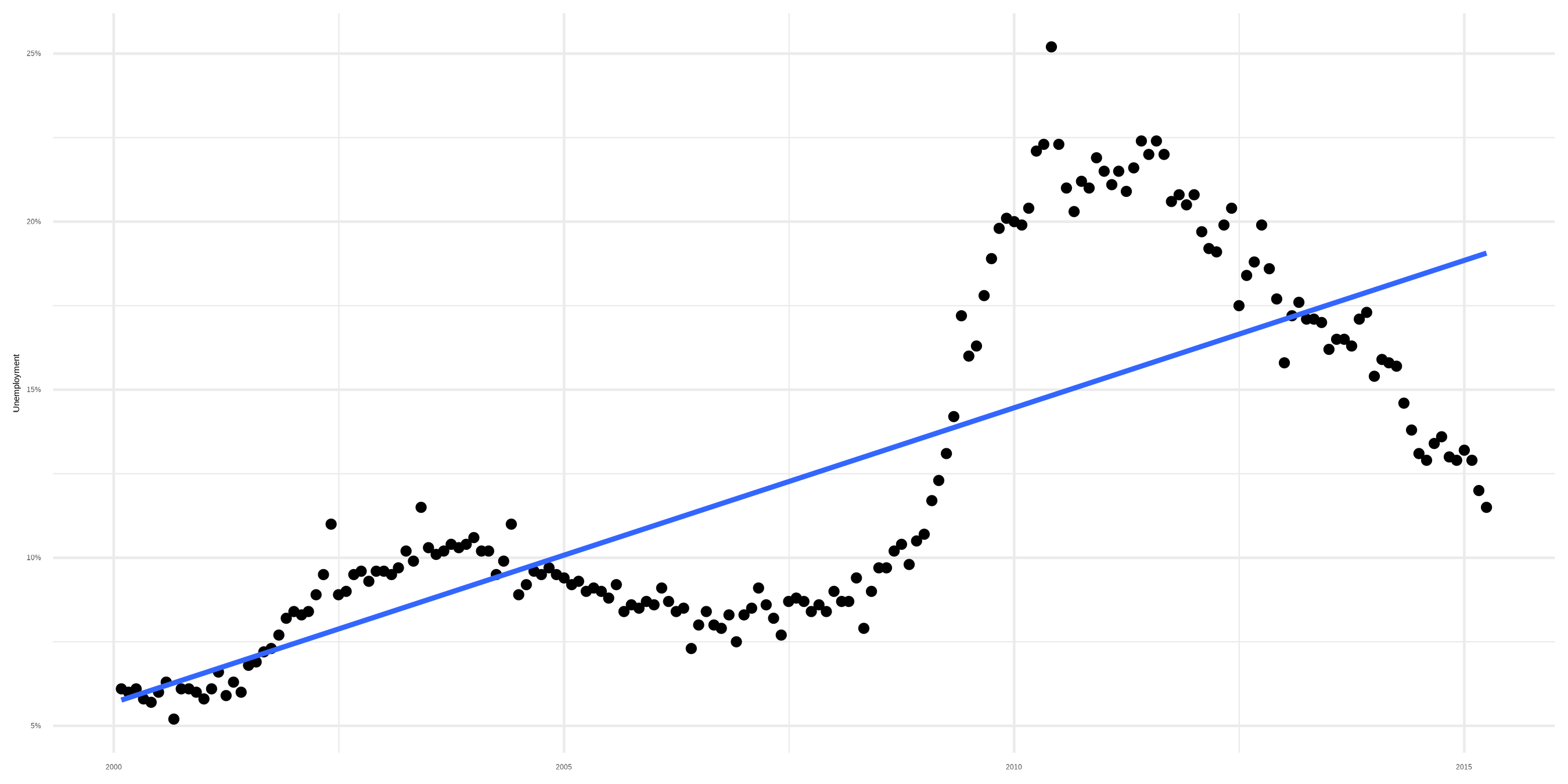
Smoother with bad linear fit
Sidenote: Smoothers
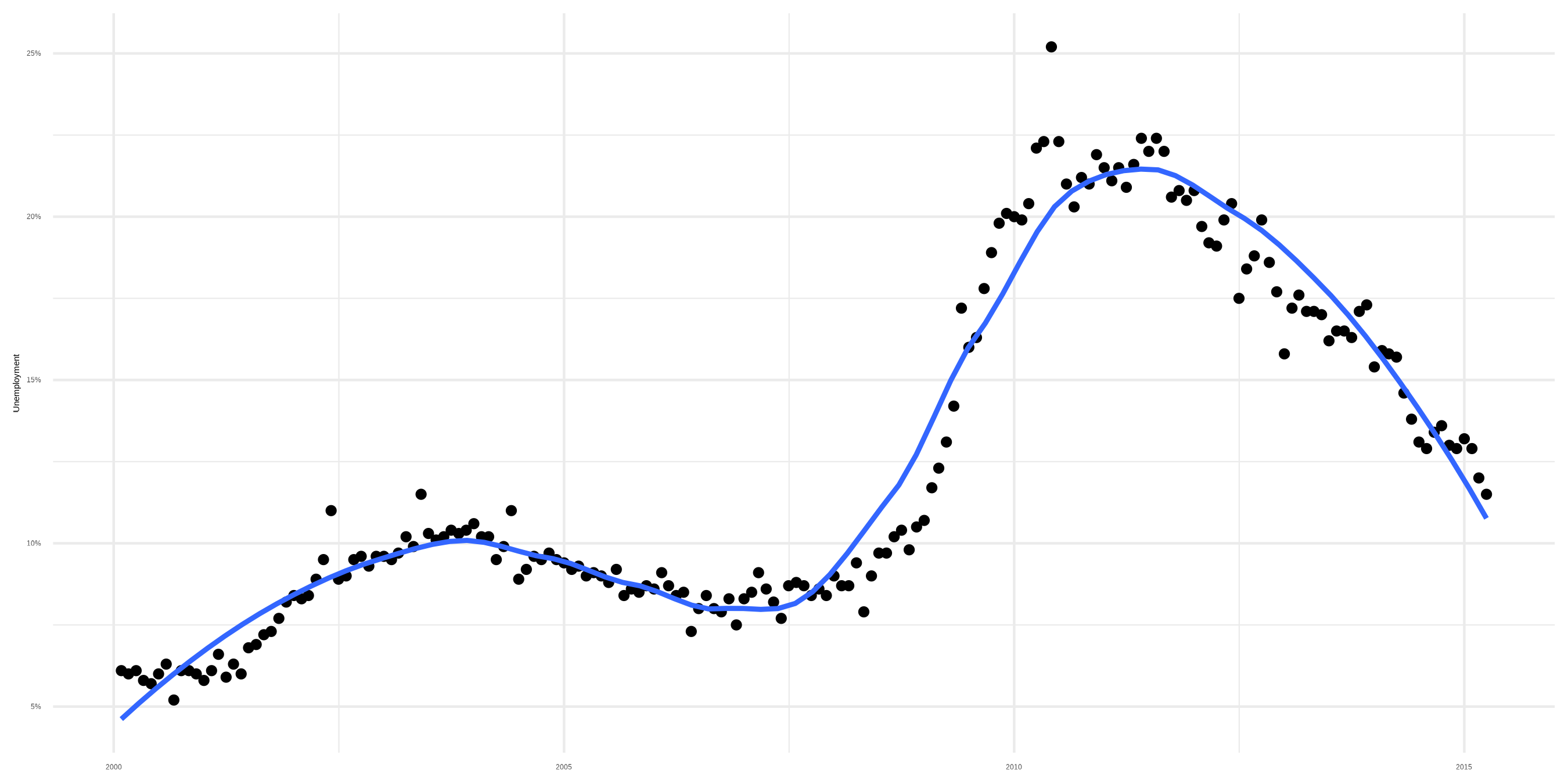
Smoother with loess fit
















